Reactive versus neoplastic – New panel to clarify clonal T-cell populations
Due to their clinical, histopathological, and molecular heterogeneity, mature T-cell neoplasms pose a major diagnostic and therapeutic challenge. Unlike the more common mature B-cell neoplasms, in which clonality is evidenced by immunoglobulin light chain restriction, most mature T-cell neoplasms lack a specific immunophenotypic signature. Wherever a suspect mature T-cell population is detected by immunophenotyping, a molecular genetic test to determine the clonality of T-cell receptor rearrangements is currently recommended for further diagnostic clarification and for differentiation from reactive changes. However, a positive finding is not a reliable criterion for malignancy in this case, since non-malignant clonal expansion of T cells can occur as an excessive immune reaction, e.g., with infections, autoimmunity, or after administration of certain drugs. In routine diagnostics, additional genetic analyses of more meaningful markers are therefore required to enable a clear distinction between benign and malignant clonal T-cell populations and thus shorten the time to diagnosis.
Next generation sequencing in patients with a clonal T-cell population
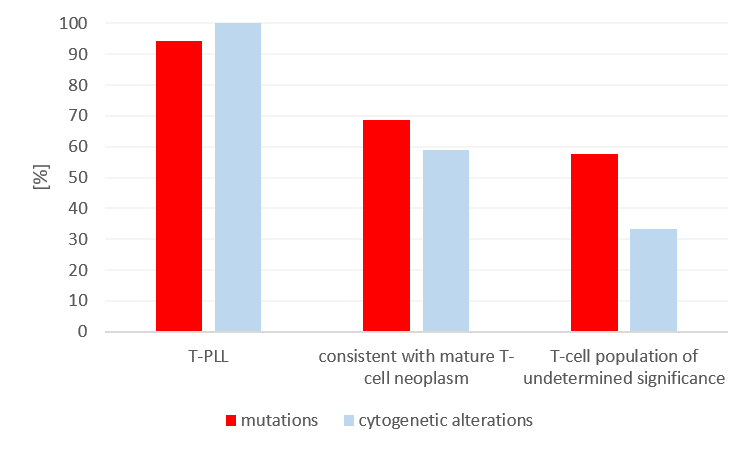
To evaluate the potential benefit of genetic analysis in patients with a suspected mature T-cell population, NGS was performed in 83 patients using a lymphatic gene panel. All patients had previously been tested using immunophenotyping, and cytogenetic data was available for 52 patients. Based on the results of immunophenotyping, 18 patients had T-PLL, 32 patients had findings consistent with another mature T-cell neoplasm (TCL), and 33 patients had a T-cell population of undetermined significance (TPUS). In all patients, it was possible to detect a clonal T-cell receptor rearrangement in the molecular genetic analysis. Due to its specific immune phenotype and its entity-defining cytogenetic changes (TRAD::TCL1A rearrangement or TRAD::MTCP1 rearrangement), T-PLL is comparatively easy to diagnose – all 18 patients showed the T-PLL-typical cytogenetic changes and 94% of the cases also had mutations. Structural changes and/or copy number changes were also detected by chromosome analysis and FISH in 59% of TCL and 33% of TPUS patients. As many as 69% of the TCL and 58% of the TPUS patients showed mutations. While mutations in JAK3, ATM, JAK1, and STAT5B primarily occurred in T-PLL, mutations in STAT3, TET2, and DNMT3A were mainly observed in TCL and TPUS. STAT3 mutations commonly occur in approximately 30-40% of T-LGLs, but they also occur in other mature T-cell neoplasms. It is worthy of note that 60% of the TPUS patients exhibited cytogenetic and/or molecular genetic markers.
In the meantime, there have been numerous publications that describe in detail the molecular landscape of the various T-cell neoplasms. Thanks to larger data sets and expanded sequencing approaches, the diagnostic and prognostic value of certain molecular changes is becoming increasingly apparent. Most notable is the high number of overlapping mutations in epigenetic modifiers in angioimmunoblastic T-cell lymphoma (AITL), which is referred to as nodal follicular T-cell lymphoma of the angioimmunoblastic type (nTFHL-AI) according to the WHO classification of 2022 (Aleggio et al. Leukemia 2022). These include TET2 (50%-80%), DNMT3A (20%-30%), and IDH2-R172 (20%-30%). The RHOA-G17V mutation is also observed in up to 70% of nTFHL-AI cases. However, none of these mutations are specific for nTFHL-AI, as they can also be observed in other entities, particularly in nodal PTCL with a TFH phenotype (nTFHL-NOS according to the new WHO classification). Cytogenetic changes with diagnostic value include ALK rearrangements in ALK-positive anaplastic large cell lymphoma (ALCL), rearrangements of DUSP22 or TP63 in ALK-negative ALCL, and ITK::SYK fusions in follicular T-cell lymphoma (nTFHL-F according to the new WHO classification).
Clinical relevance of molecular genetic changes
While an effective targeted therapy with brentuximab vedotin is only available for CD30-positive anaplastic large cell lymphoma (ALCL), clinical trials are primarily testing histone deacetylase inhibitors (HDACi) at present. In particular, R/R T-cell neoplasms with the TFH phenotype showed a significantly better response compared to T-cell neoplasms without the TFH phenotype (HR: 0.322; p = 0.009). They also exhibited a typical mutation pattern from TET2 and/or DNMT3A and/or RHOA mutations more frequently (83% vs 40%; p = 0.034) (Ghione et al. Blood Adv 2020).
Based on this data and the clinical relevance, an extended genetic investigation of clonal T-cell populations looks promising. We have therefore expanded our range of tests and are offering a new T-cell-specific panel in molecular genetics, including the genes TET2, DNMT3A, RHOA-G17V, IDH2-R172, CD28, FYN, PLCG1, and VAV1 (see our current test order as well as our digital order entry system (only available in German language)).
The author

»Do you have questions regarding this article or do you need further information? Please send me an e-mail.«
Dr. rer nat. Janine Müller