IPSS-M: Molecular International Prognostic Scoring System
Risk scoring for patients with myelodysplastic neoplasms (MDS) has been based on clinical variables and cytogenetic aberrations as part of the Revised-International Prognostic Scoring System (IPSS-R). A decade of molecular genetic analysis emphasized the impact of mutations (Bejar et al. NEJM 2011, Papaemmanuil et al. Blood 2013, Haferlach et al. Leukemia 2014). The Molecular International Prognostic Scoring System (IPSS-M) now takes mutations in 31 genes in account, as well as cytogenetics, bone marrow blasts, hemoglobin and platelet count (Bernard et al. NEJM Evid 2022).
What is the IPSS-M?
An international research team (The International Working Group for the Prognosis of MDS) analyzed 2,957 MDS patients and developed an improved prognostic score using the following parameters:
- Hemoglobin
- Platelet counts
- Bone marrow blasts
- IPSS-R cytogenetic risk category
- Molecular genetic information on 31 genes
The authors particularly emphasize the importance of FLT3 alterations (TKD or ITD) and KMT2A-PTD (MLLPTD). While very rare in MDS, they have a highly adverse impact on the prognosis. For TP53, the status "multihit" plays a significant role; this is referred to as the presence of two or more mutations, a mutation and a deletion (del(17p)) or a mutation with a copy-neutral loss of heterozygosity (cnLOH).There are also new results regarding SF3B1. The positive prognostic significance is lost, if present with a del(5q) or a mutation in one of the following genes: BCOR, BCORL1, NRAS, RUNX1, SRSF2, or STAG2.
How can the IPSS-M be calculated?
The following is needed in addition to the blood counts:
- Cytomorphology (bone marrow blast count)
- Cytogenetics/chromosome analysis
- The IPSS-M panel according to Bernard et al. in molecular genetics
The result of the IPSS-M is a number, which is classified into six risk categories (see Fig. 1):
- Very Low (VL)
- Low (L)
- Moderate Low (ML)
- Moderate High (MH)
- High (H)
- Very High (VH)
Fig 1. Shows the classification of numerical values to the risk groups and their relative frequency in MDS (Bernard et al. NEJM Evidence 2022).
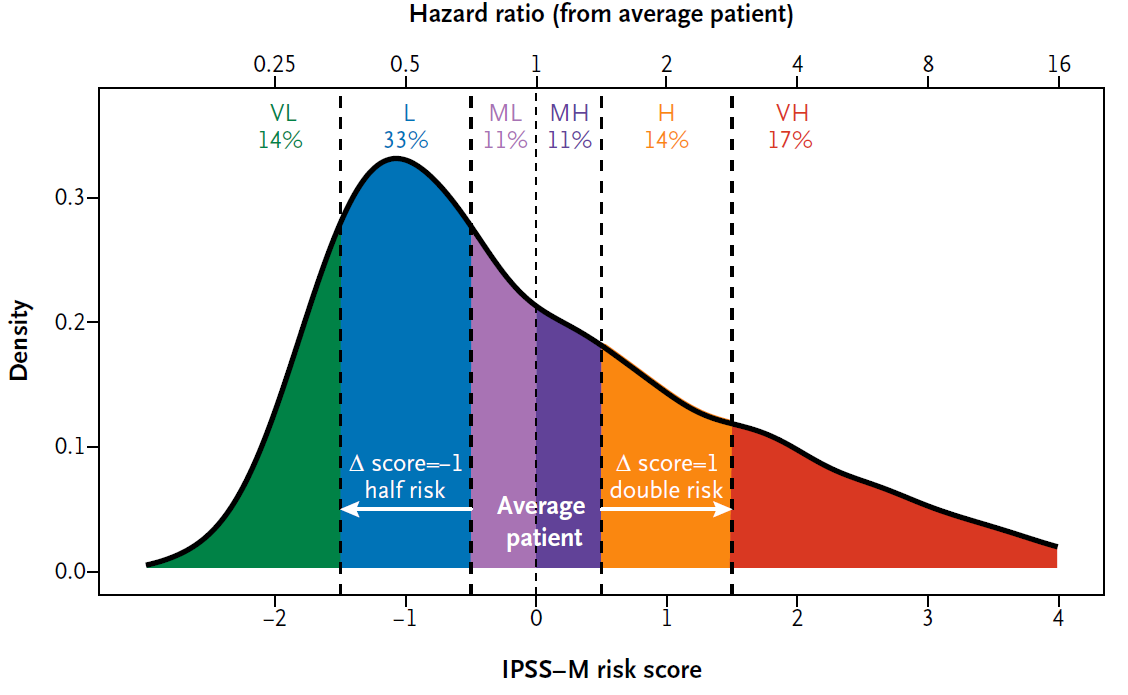
In the publication, the risk groups show strong prognostic separation in terms of overall survival, leukemia-free survival, and AML transformation.
The IPSS-M can be calculated on the MDS Foundation website:
A few practical tips for use:
- At the end of our report, you will find the table Conducted
analysis. Enter a tier 1 and 2 type mutation as “Mutated.” - The original gene name MLL is used in the
WebTool. We use the current name KMT2A. - TP53: You can find the number
of mutations in the Mutations Changes. In addition, the “Maximum TP53VAF”
must be entered. This can be found in the VAF column in the same table. In
the future, we will also provide information on the “Loss of
heterozygosity”
We are currently updating our reports in accordance with the IPSS-M. If you send us blood counts and ask for cytomorphology, cytogenetics and molecular genetic diagnostics, we will soon also be able to provide you with the IPSS-M as part of our integrated report.
The author

»Do you have questions regarding this article or do you need further information about the IPSS-M? Please send me an e-mail.««
Dr. rer. nat. Constance Bär
Biologist
Head of Sequencing