Mantle Cell Lymphoma (MCL)
- Method:
- Anticoagulant:
- Recommendation:
- Method:Cytomorphology
- Anticoagulant:EDTA
- Recommendation:obligatory
- Method:Immunophenotyping
- Anticoagulant:EDTA or Heparin
- Recommendation:obligatory
- Method:Chromosome analysis
- Anticoagulant:Heparin
- Recommendation:facultative
- Method:FISH
- Anticoagulant:EDTA or Heparin
- Recommendation:obligatory
- Method:Molecular genetics
- Anticoagulant:EDTA or Heparin
- Recommendation:facultative
Based on the current guidelines and the current state of research, there are different diagnostic recommendations for patients with mantle cell lymphoma. We have summarized the most important information on classification and diagnostic methods at the MLL. In addition, we provide further links on prognosis and therapy in mantle cell lymphoma, so that you can inform yourself in more detail.
Mantle cell lymphoma: Classification
Mantle cell lymphoma (MCL) is classically a mature B-cell neoplasia with indolent to aggressive courses. Characteristic is the chromosomal translocation t(11;14)(q13;q32) (IGH::CCND1) with overexpression of cyclin D1, which is present in more than 95% of cases. According to WHO, mantle cell lymphoma (MCL) is subdivided into the following subtypes depending on clinicopathologic features and underlying pathogenic pathways, as shown in Figure 1 (WHO 2022):
- In situ mantle cell neoplasm (ISMCN)
- Mantle cell lymphoma (MCL)
- Leukaemic non-nodal mantle
cell lymphoma (nnMCL)
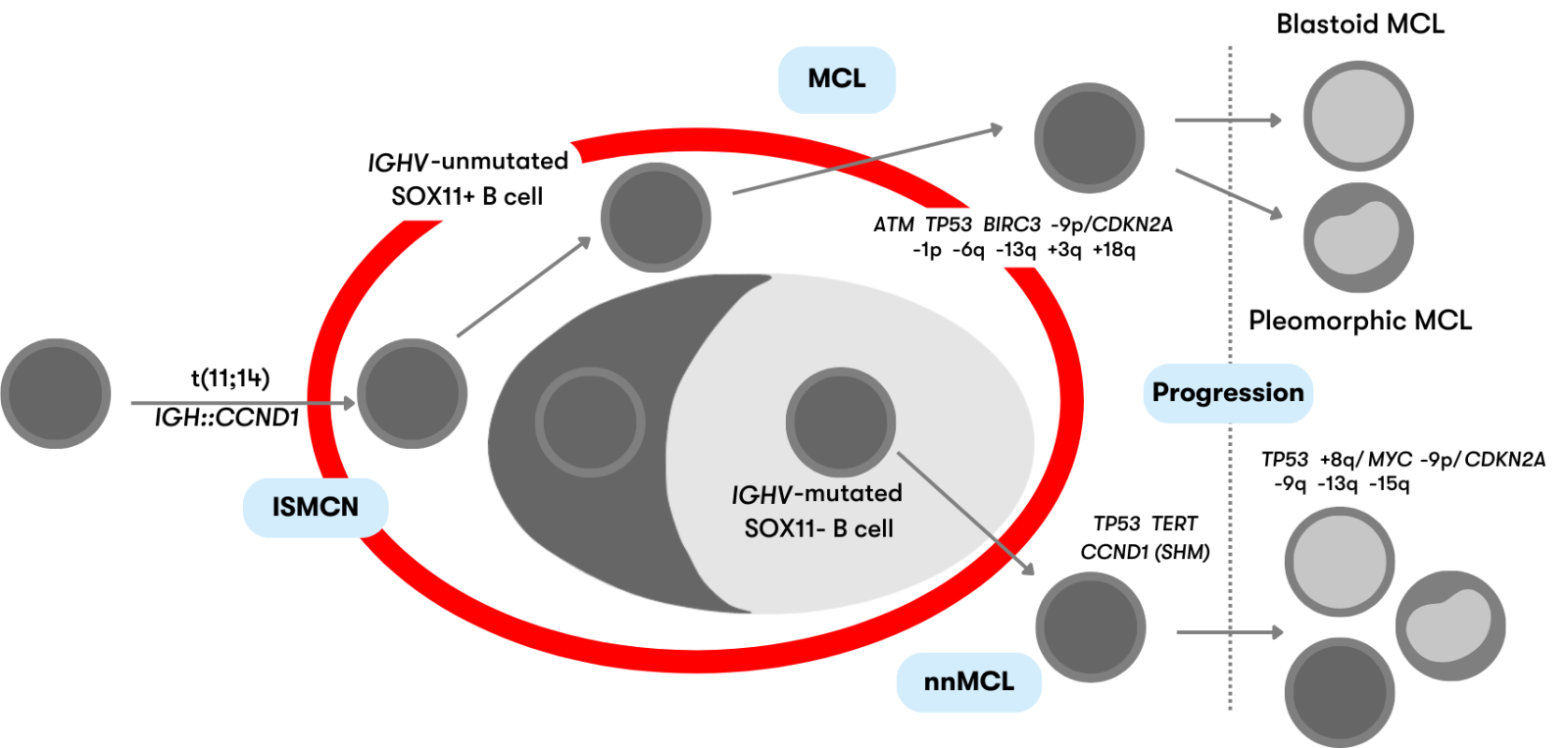
Figure 1: Schematic representation of the defined mantle cell lymphoma subtypes (modeled after WHO 2022)
ISMCN are mostly indolent and usually incidental findings in lymph nodes. B cells are present in the mantle zone of the lymphoid follicle and show CCND1 overexpression due to the IGH::CCND1 fusion. ISMCN rarely develop into MCL (<10%) (WHO 2022).
Classic MCL derives from naive-like mature B cells. The IGH::CCND1 fusion is present in >95% of MCL. MCL typically show high SOX11 expression, mutations of the genes ATM and TP53, copy number gains of 3q, and copy number losses of 13q and 1p. Patients usually have generalized lymphadenopathy and an unfavorable course (Nadeu et al. 2020, WHO 2022).
IGH::CCND1 fusion is also present in nnMCL arising from memory-like B cells. They are characterized by involvement of peripheral blood, bone marrow, and spleen with little or no lymphadenopathy and a usually asymptomatic clinical presentation. nnMCL typically show no or low SOX11 expression and fewer genetic aberrations than classic MCL (Nadeu et al. 2020, WHO 2022). SOX11-negative MCL are the much rarer variant and account for approximately 14%-32% of MCL (Cheah et al. 2016).
Mantle cell lymphoma: Diagnostic methods
Mantle cell lymphoma: Prognosis
Prognostically unfavorable markers include TP53 deletions and mutations, CDKN2A deletions, NOTCH1/2 mutations, and Ki-67. TP53 mutations have an even worse impact on prognosis compared to TP53 deletions (Rubio-Moscardo et al. 2005, Salaverria et al. 2007, Sander et al. 2008, Cheah et al. 2016, Hoster et al. 2016, Eskelund et al. 2017). Age, poor general health, advanced disease stage (Ann Arbor stage III or IV), splenomegaly and anemia, serum levels of β2-microgloblulin and LDH, blastoid cytology, extranodal presentation, and constitutional symptoms are among important clinical and serologic factors associated with an unfavorable course (Silkenstedt & Dreyling 2023).
The MIPI (MCL International Prognostic Index) is established as a clinical risk score. In combination, the MIPI and Ki-67 index form the MIPI-c (combined MIPI) for risk assessment (Hoster et al. 2008, Hoster et al. 2016). Click here for the prognosis calculation of the MIPI and the MIPI/MIPI-c.
Mantle cell lymphoma: Therapy
The choice of therapy should be made in the context of the mutation status (Martin et al. 2017). According to the current Onkopedia guideline on mantle cell lymphoma, which also provides detailed information on therapeutic strategies, patients with MCL should be treated in the context of clinical trials whenever possible. This is especially true for patients with a high burden of copy number alterations (Martin 2018).
Mantle cell lymphoma: Recommendation
According to the current Onkopedia guideline on mantle cell lymphoma, besides the collection of clinical and laboratory parameters from peripheral blood, cytological and histological examination of bone marrow is recommended, and in case of leukemic course, immunophenotyping of surface markers from peripheral blood is also recommended.
Status: September 2023