New possibilities for more sensitive investigation of measurable residual disease (MRD)
Measurable residual disease (MRD) is an important biomarker in acute myeloid leukemia (AML), an indicator of prognosis and response to therapy. In this article, we explain how mutations in AML and other diseases can be detected with a sensitivity of 0.1%. From July 2024, we will offer a corresponding panel and thus expand our portfolio for follow-up examinations.
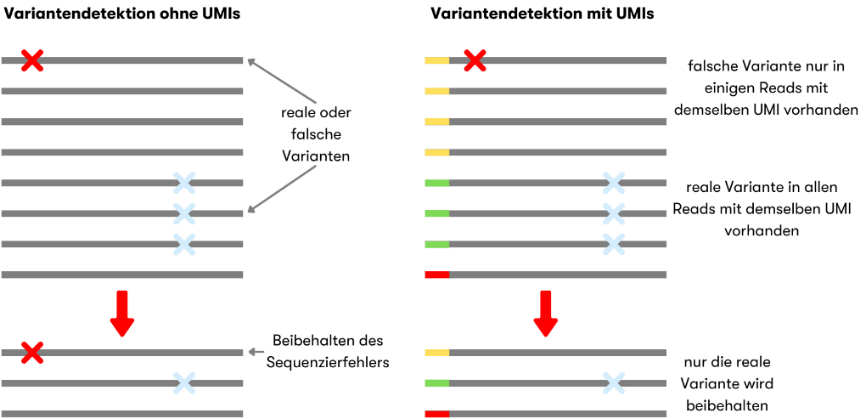
For fusion genes (e.g. RUNX1::RUNX1T1) or typical NPM1 changes, sensitive quantitative real-time (qRT)PCRs or digital droplet (dd)PCRs have been available for more than ten years. Genes such as RUNX1 or TP53, on the other hand, have a large number of different mutations. Here, sequencing of the complete gene is the method of choice. The sensitivity of Next Generation Sequencing (NGS) is limited by the natural error rate of the enzymes used, among other things. The latter can be corrected with a trick. Each individual DNA molecule is provided with a short individual sequence, a unique molecular identifier (UMI), at the beginning of the preparation. In this way, all the results of an original piece of DNA can be added together bioinformatically. If an error has crept in, it is recognized and eliminated (Figure 1). Such sequencing provides data with a low error rate. In combination with a higher sequencing depth, even changes of 0.1 % can be reliably detected.
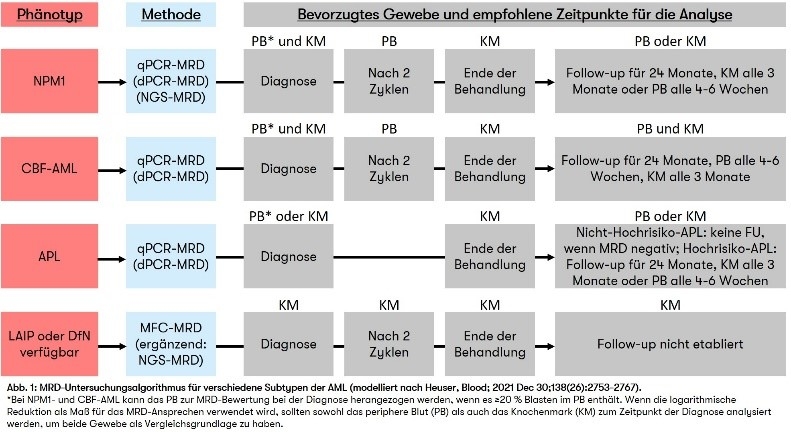
The new technical possibilities are also included in the guidelines of the European LeukemiaNet (ELN) for the standardization and harmonization of MRD (Heuser, Blood; 2021 Dec 30;138(26):2753-2767). In addition to the established methods (ddPCR and qRTPCR), UMI-supported NGS is also listed in the guidelines. In our UMI panel we cover all genes of the ELN recommendation. Specifically for AML, the ELN guidelines define clinically relevant time points for MRD testing, which are intended to provide assistance in everyday life and can contribute to further standardization (Figure 2).
How to request a follow-up examination:
- When an alteration is diagnosed for the first time, we indicate in the appendix of the findings whether and with what sensitivity each genetic alteration can be used as a marker for disease progression.
- As each patient has their own set of markers, we will create a personalized MRD approach using one or more of the above methods upon resubmission. We always use the most sensitive method available to us.
- You should generally receive the results within 10 working days.
- In addition to the molecular genetic methods mentioned above, immunophenotyping can also make a valuable contribution to MRD testing.
We use these methods not only for AML, but also for other diseases, so that as many patients as possible can benefit from optimal MRD testing.
Heuser M et al. 2021 Update on MRD in acute myeloid leukemia: a consensus document from the European LeukemiaNet MRD Working Party 2021; Blood; 2021 Dec 30;138(26):2753-2767

"If you have any further questions on this topic, please contact us!"
Dr. rer. nat. Constance Bär
Biologist
Head of Sequencing