62. ASH Annual Meeting & Exposition - a follow-up report
Also in 2020, this time as a virtual congress, the MLL was well represented at the 62nd Annual Meeting & Exposition of the American Society of Hematology (ASH) with its own presentations and in collaborations with others. Overall, the MLL and its team presented four seminars, 9 posters and one short lecture.
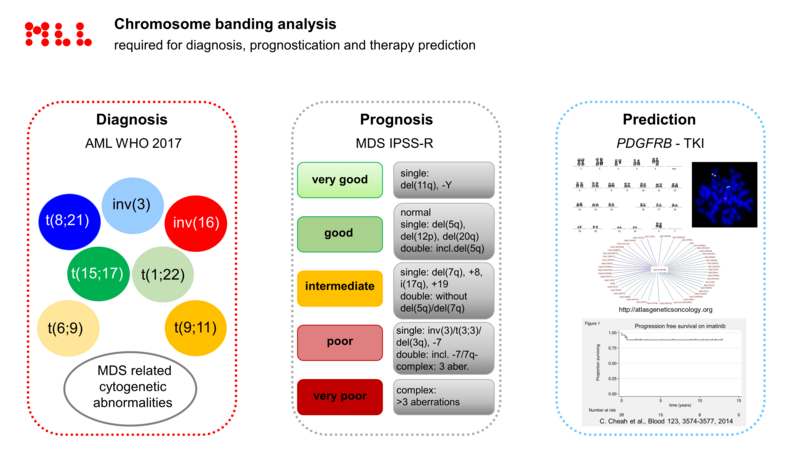
The focus was on the more precise characterization and classification of leukemia and lymphoma as well as the application of new methods extending to genome sequencing and artificial intelligence, both in research and in routine diagnostics.
A total of 12 scientists from the MLL presented the results from the research projects. At a cytomorphology seminar, Dr. C. Pohlkamp presented recent results from the artificial intelligence-assisted differentiation of peripheral blood counts (a project in collaboration with Amazon Web Services). Correct predictions for individual types of benign and malignant cells in peripheral blood could be made at a rate of 92% with the tested algorithm. The tools developed for this can be used in a cloud-based manner and are currently under prospective testing (BELUGA study) parallel to the gold standard of peripheral blood count differentiation by medical technicians and hematologists.
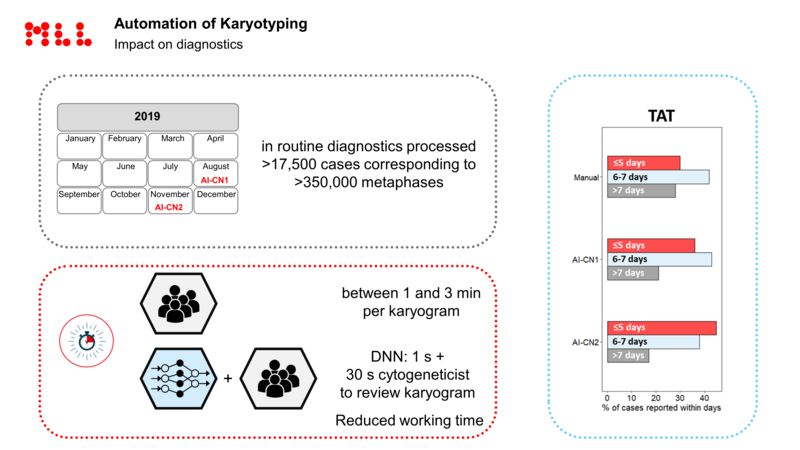
Prof. C. Haferlach then presented the current state of affairs for classic chromosomal analysis: The MLL has now succeeded in developing algorithms together with MetaSystems that can assist medical technicians and biologists with karyotyping. Well over 20,000 patients have already been processed in routine diagnostics using artificial intelligence algorithms, and this has led to a significant reduction in diagnostic times for chromosome analysis, sometimes by as much as two days compared to the previous workflow duration.
Panel sequencing with next-generation sequencing (NGS) is playing an increasingly important role in molecular genetics. For unclarified issues and excluding MDS in particular, clear benefits become evident for clinical decision-making as well as for further diagnostics and therapy selection. In her presentation, Dr. C. Bär, was able to show that with unclear constellations such as cytopenias, the analysis of a panel of genes frequently mutated in myeloid neoplasias leads to a significant improvement in the clinical diagnostic algorithms and to a reduction of differential diagnoses. For both the patient and the hematologist, this results in a reduced time to a definitive or exclusion diagnosis, while the otherwise commonly practiced multiple repeated punctures for the same clinical blood count constellation can also be reduced.
Together with ASH, the MLL has over the past two years conducted a project, whose results were presented by Mr N. Nadarajah at the ASH meeting. This involved the harmonization of molecular genetic findings, which, under the leadership of the MLL and through joint data analysis by six different laboratories in Europe and the USA, has led to a jointly agreed assessment of variants in genes that are altered in hematological diseases. The evaluation algorithms and the laboratory workflow were discussed and compared between laboratories. The result for diagnosing hematological neoplasias was published on the ASH homepage in October 2020 and is now being further developed and expanded by us.
The aim of the MLL’s scientific activities will therefore continue to improve the diagnostics for our patients, to share and make the data available for scientific publications and collaborations, and to advance new technological options such as NGS and the use of artificial intelligence to support routine laboratory diagnostics.
In the following we have summarized all the ASH projects involving MLL:
The author

»Do you have questions regarding this article or do you need further information? Please send me an e-mail.«
Prof. Dr. med. Dr. phil. Torsten Haferlach
Executive management
Internist, Hematologist and Oncologist
Deputy Head of Cytomorphology
torsten.haferlach@mll.com