Our First Whole Genome Sequencing Report
Whole Genome Sequencing in Routine Diagnostics
Whole genome
sequencing (WGS) detects a wide range of genetic alterations in a single analysis: from
point mutations to structural alterations to chromosomal copy number
alterations. Thanks to our 5,000
genome project,
we have been able to acquire a lot of experience in the laboratory in
recent years, but primarily in the processing and analysis of the
large-scale and extensive data as well. We at MLL have now taken the
next step and bundled this experience, transferring it to routine
diagnostics and designing a corresponding whole genome sequencing report
for it.
A Report Layout Designed Specifically for Whole Genome Sequencing
The focus of the new layout here is the clear presentation of the
results in the report, without diminishing the extent of the data
analyzed. In our standard evaluation pipeline, single nucleotide
variants (SNVs), structural variants (SVs), copy number variations
(CNVs), and copy-neutral loss of heterozygosity (CN-LOH) are
investigated and reported. Along with a brief description of the changes
found, the findings include a graphical presentation of all results as a
Circos plot (see figure).
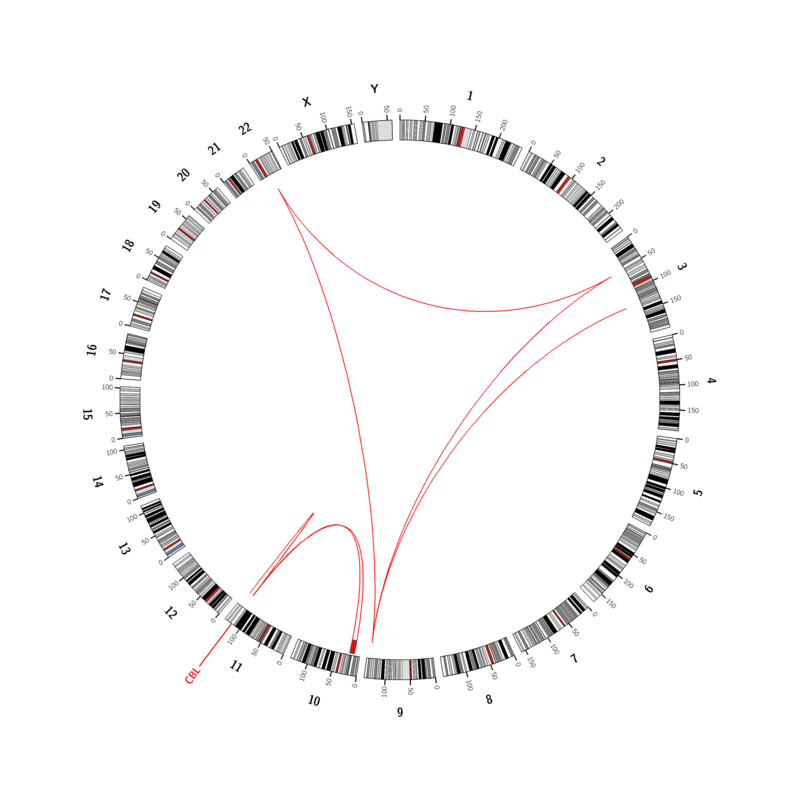
Our first clinical report of whole genome sequencing involved CML in blast crisis, which, besides a complex 3-way translocation between chromosomes 3, 9, and 22 leading to the BCR-ABL1 fusion transcript, revealed a second additional KMT2A-MLLT10 translocation between chromosomes 10 and 11. In addition, a mutation (SNV) in CBL and the loss of chromosomal material from chromosome 10 was able to be identified.
In our Circos plot, the chromosomes are listed in circular form, translocations are represented by red connecting lines, gains (blue) and losses (red) are marked inside the chromosome circle, and the gene with a mutation is labeled at its position in the genome. This graphical representation of the results is intended to give the treating physician a quick overview of the genome-wide alterations. Naturally – as usual – this report also contains the relevant alterations in text form, tables with detailed information, and the resulting interpretation with regard to diagnosis and prognosis.
The First Clinical Report Has Been Sent Out
We are pleased to announce that, in mid-June, we were able to send out
the first clinical report on whole genome sequencing in the new layout
with the added graphic. The additional information that whole genome
sequencing can bring will become clear in the future. It is therefore
all the more gratifying that for complex cases and diseases such as Multiple myeloma and acute
lymphatic leukemia (ALL),
we are already able to contribute to the further development of
diagnostic methods, collect therapy-controlling findings, and clearly
present the results in a disease-relevant way through whole genome
sequencing and transcriptome analyses.
The author

»Do you have questions regarding this article or do you need further information? Please send me an e-mail.«
Dr. rer. nat. Manja Meggendorfer, MBA
Biologist, Dipl.
Head of Molecular genetics
Head of Research and Development
manja.meggendorfer@mll.com