Clonal hematopoiesis of indeterminate potential - CHIP in Hematology
- Method:
- Anticoagulant:
- Recommendation:
- Method:Cytomorphology
- Anticoagulant:EDTA
- Recommendation:obligatory*
- Method:Immunophenotyping
- Anticoagulant:
- Recommendation:no
- Method:Chromosome analysis
- Anticoagulant:Heparin
- Recommendation:no
- Method:FISH
- Anticoagulant:EDTA or Heparin
- Recommendation:no
- Method:Molecular genetics
- Anticoagulant:EDTA or Heparin
- Recommendation:obligatory
*in case of positive molecular genetics
Based on the current guidelines and the current state of research, different diagnostic recommendations arise for patients with clonal hematopoiesis of indeterminate potential. We have summarized the most important information on classification and diagnostic methods at MLL. In addition, we provide further links and literature on clonal hematopoiesis of undetermined potential.
CHIP in hematology: Classification
Clonal hematopoiesis of indeterminate potential (CHIP) is considered a myeloid precursor lesion according to the WHO 2022 classification.
CHIP definition of WHO 2022 (WHO 2022)
- Detection of one or more somatic mutations with variant allele frequency (VAF) ≥2% (≥4% for X-linked gene mutations in males) in the DNA of blood or bone marrow cells involving selected genes (see Molecular Genetics, Table 2)
- Absence of unexplained cytopenias
- Absence of diagnostic criteria for defined myeloid neoplasms
CHIP is rare in individuals under 40 years of age and increases steadily after 65 years. In older individuals, it affects between 10-40%, with prevalence also depending on the sensitivity of the diagnostic sequencing method (Haferlach & Heuser 2022, WHO 2022).
Individuals with CHIP show an increased risk of developing hematologic neoplasia. The risk for other diseases, such as cardiovascular disease, is also increased in the presence of CHIP. You can learn more about CHIP in cardiology here.
Differentiation of CHIP from CCUS and MDS
When clonality of hematopoiesis is detected, it is necessary to differentiate CHIP from CCUS (clonal cytopenia of undetermined significance) and full-blown myeloid neoplasms, especially myelodysplastic neoplasms (MDS).
If clonal hematopoiesis is accompanied by cytopenia of unclear cause, this is referred to as CCUS. If cytomorphological signs of dysplasia are also present, the diagnostic criteria of MDS are fulfilled.
Table 1: Differentiation of CHIP and CCUS from MDS (Hoermann 2022 [2])
|
|
CHIP |
CCUS |
Low risk MDS |
High risk MDS |
|
Clonality |
+ |
+ |
+ |
+ |
|
Dysplasia |
- |
- |
+ |
+ |
|
Cytopenia |
- |
+ |
+ |
+ |
|
BM Blasts |
<5% |
<5% |
<5% |
5-20% |
|
Cytogenetic aberrations |
+/- |
+/- |
+ |
++ |
|
Molecular aberrations |
+ |
+ |
++ |
+++ |
|
Risk of progression |
+ |
++ |
++ |
+++ |
CHIP in hematology: Diagnostic methods and their relevance
CHIP in hematology: Prognosis
The transformation rate in the presence of a CHIP is 0.17-0.22% per year (Haferlach & Heuser 2022). Large clones or the presence of multiple mutations increase the risk of progression to myeloid neoplasia. In particular, mutations in TP53, U2AF1, SRSF2, IDH1, IDH2, SF3B1, and ASXL1 increase the risk of progression. In addition, the simultaneous presence of mosaic clonal alterations (detected by microarray or WGS studies) is considered an independent risk factor for the development of leukemia (Haferlach & Heuser 2022, WHO 2022, Hoermann 2022 [1], Hoermann 2022 [2]). In the age group of ≥80 years an association of CHIP with a lower survival probability has been described especially in the presence of ≥2 mutations. In this age group, in addition to specific mutation patterns, a mutation allele frequency of ≥9.6% has been identified as a risk factor for the development of myeloid neoplasia (Rossi et al. 2021).
The Clonal Hematopoiesis Risk Score (CHRS) was established in 2023 as a prognostic risk score for the development of a haematological neoplasia. This incorporates various variables that lead to classification into low, intermediate and high risk groups. The sole presence of a DNMT3A mutation is considered a prognostically favorable factor in CHRS. In contrast, the following parameters are considered to be prognostically unfavorable factors (Weeks et al. 2023):
- defined high-risk mutation (SRSF2, SF3B1, ZRSR2, IDH1, IDH2, FLT3, RUNX1, JAK2 and TP53)
- Detection of multiple mutations
- Clone size of at least 20% variant allele frequency (VAF)
- Red cell distribution width (RDW) of ≥15%
- Mean corpuscular volume (MCV) of ≥100 fl
- Presence of cytopenia (CCUS vs. CHIP)
- Age ≥65 years
The nature of CHIP alterations influences the resulting myeloid (M-CHIP) or lymphoid (L-CHIP) hematologic disease (Niroula et al. 2021, Haferlach & Heuser 2022, Hoermann 2022 [1]). Mutations of TP53 and IDH1/2 and spliceosome genes also increase the risk of developing AML (Abelson et al. 2018, Desai et al. 2018) CHIP is associated with an increased risk of all-cause mortality, atherosclerotic cardiovascular disease, and autoimmune disease (WHO 2022). CHIP is also considered a risk factor for advanced chronic obstructive pulmonary disease (COPD) and the development of various other non-hematologic diseases (Haferlach & Heuser 2022). A high number of genetic alterations significantly increases the likelihood of dying from hematologic neoplasia or cardiovascular disease (Saiki et al. 2021). See also CHIP in Cardiology.
CHIP as a risk factor for the development of therapy-associated myeloid neoplasms
CHIP clones can gain a selective survival advantage and expand under hematologic stress, which can result from cytotoxic therapy, radiation, or stem cell transplantation, among others. In particular, TP53 and PPM1D mutant clones have been associated with the development of therapy-associated neoplasms following cytotoxic therapy (Wong et al. 2015, Coombs et al. 2017, Hsu et al. 2018, Kahn et al. 2018, Wong et al. 2018, Ortmann et al. 2019, Hoermann 2022 [1]).
CHIP in the context of autologous and allogeneic stem cell transplantation
A few studies have described an increased risk of developing therapy-associated myeloid neoplasia after autologous stem cell transplantation (Gibson et al. 2017). However, according to current knowledge, there is insufficient evidence to use CHIP in the context of autologous stem cell transplantation to make treatment decisions.
In allogeneic stem cell transplantation, donor CHIP status did not affect overall survival of recipients in a first study. In the context of CHIP-positive stem cell donors, an increased incidence of chronic graft-versus-host disease was observed, which was associated with a lower rate of relapse or progression (Frick et al. 2019). However, there are case reports of the occurrence of donor cell leukemia after allogeneic stem cell transplantation in the context of CHIP-positive older donors (Gondek et al. 2016, Frick et al. 2019).
CHIP in hematology: Recommendation
Also, in view of the lack of therapeutic intervention options, screening for the presence of CHIP in individuals with normal blood counts is not currently recommended from a hematologic perspective. Often, the detection of clonal hematopoiesis is an incidental finding. If the blood count is normal, patients with CHIP should have a blood count including differential blood count at regular intervals (initially after 3 months, later every 12 months) to detect possible progression. If the patient has unexplained peripheral cytopenia (CCUS), a bone marrow aspiration and differential blood count is recommended initially at 1, 2, and 3 months and subsequently every 3 months (Heuser et al. 2016). A recommendation on the diagnostic algorithm for CHIP is also given by Haferlach & Heuser 2022:
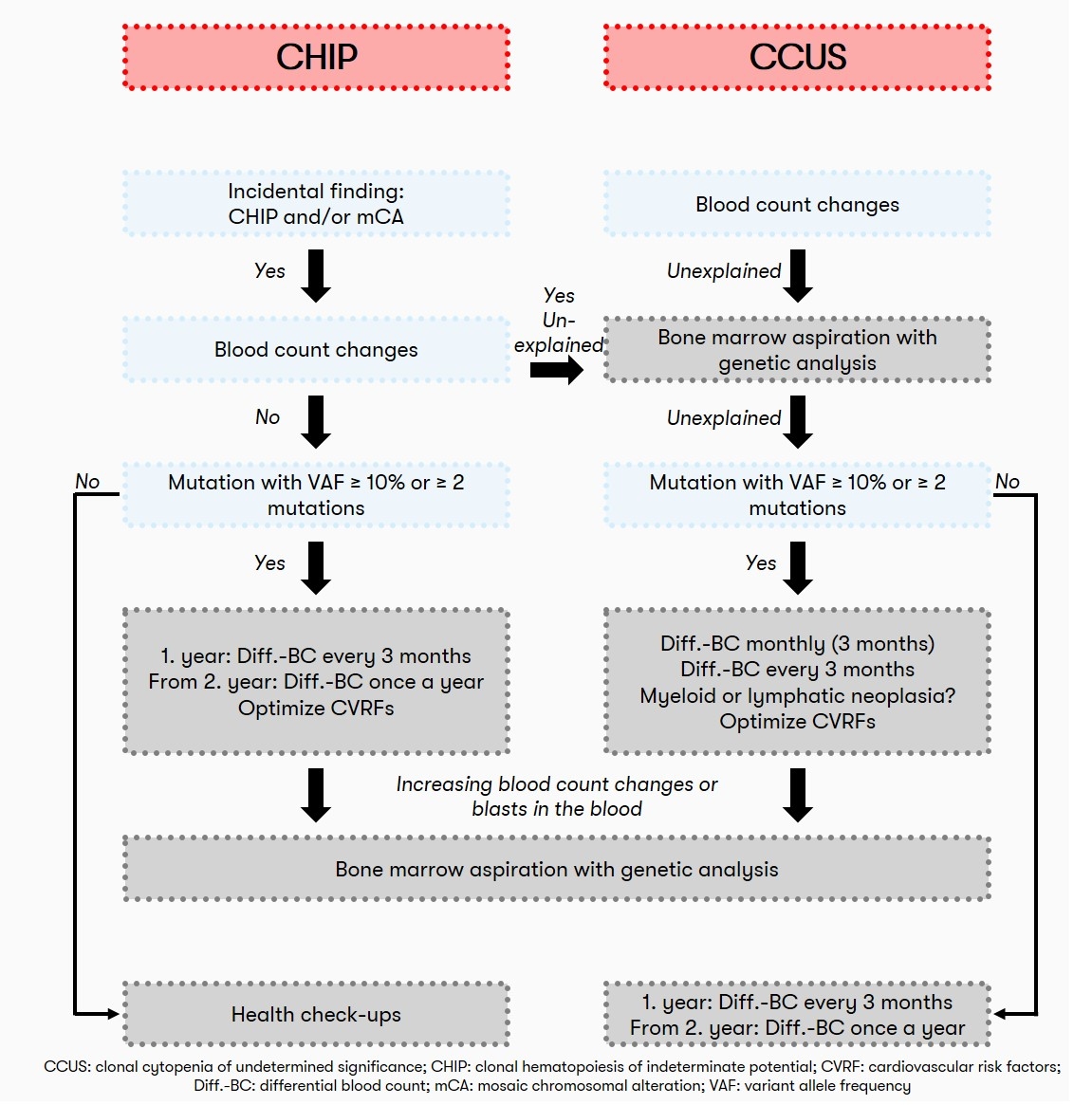
Status: November 2023