Clonal cytopenia of undetermined significance (CCUS)
- Method:
- Anticoagulant:
- Recommendation:
- Method:Cytomorphology
- Anticoagulant:EDTA
- Recommendation:obligatory
- Method:Immunophenotyping
- Anticoagulant:
- Recommendation:no
- Method:Chromosome analysis
- Anticoagulant:Heparin
- Recommendation:obligatory
- Method:FISH
- Anticoagulant:EDTA or Heparin
- Recommendation:facultative
- Method:Molecular genetics
- Anticoagulant:EDTA or Heparin
- Recommendation:obligatory
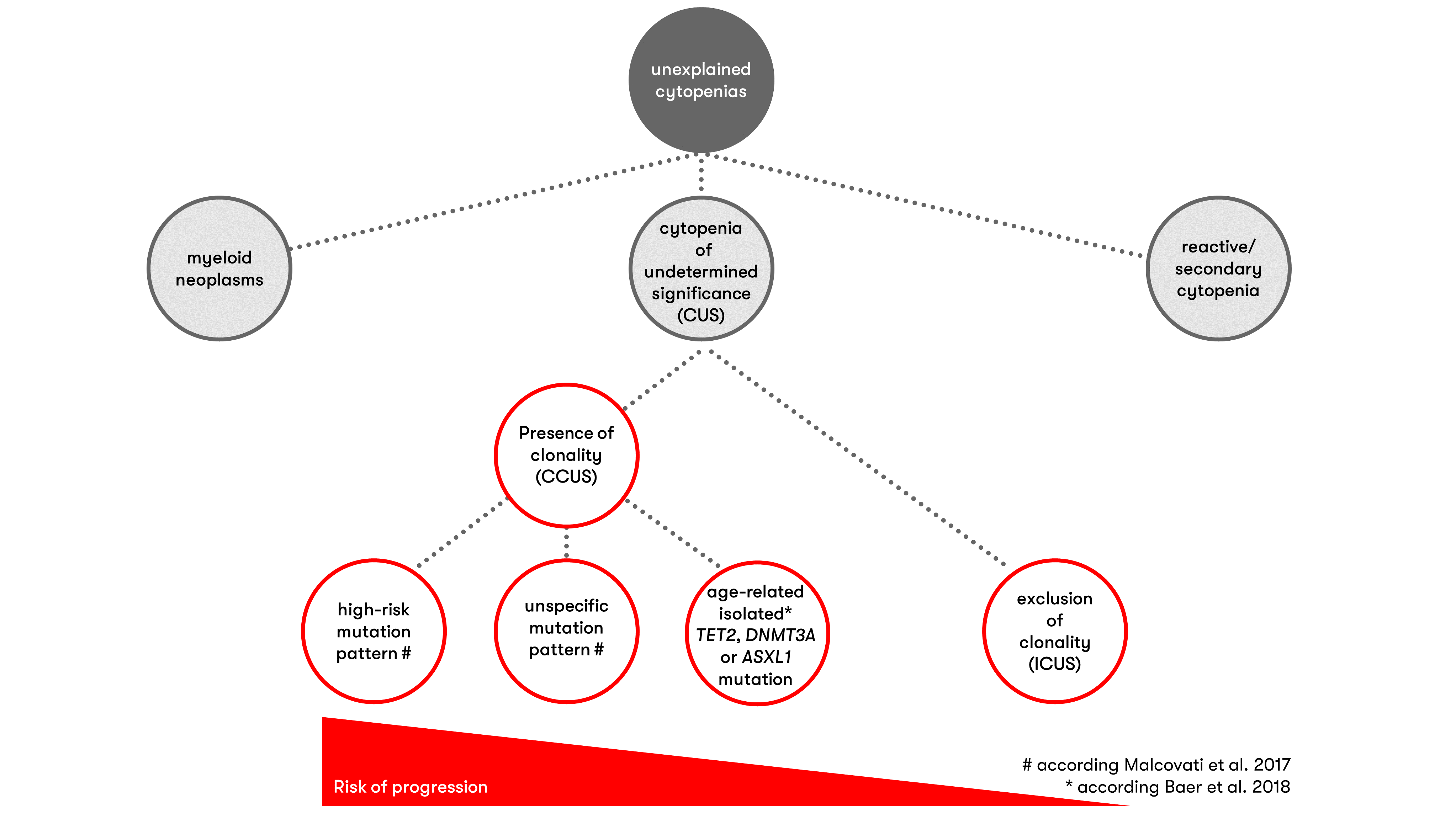
Unclear, persistent cytopenias are a frequent clinical presentation in elderly patients. In approximately one third of cases, clonal hematopoiesis is detectable in addition to cytopenia - thus fulfilling the diagnostic criteria for the diagnosis of clonal cytopenia of undetermined significance (CCUS). We provide the most important information on the classification, diagnosis and prognosis of clonal cytopenia of undetermined significance for you.
Classification of clonal cytopenia of undetermined significance
With the 5th edition, clonal cytopenia of undetermined significance is listed for the first time in the WHO classification, and it is included among the myeloid precursor diseases (WHO 2022).
Diagnostic criteria (WHO 2022)
- Evidence of clonal hematopoiesis*
- One or more persistent (>4 months) cytopenia(s) that cannot be otherwise explained
- Diagnostic criteria for defined myeloid neoplasms are not met based on bone marrow examination
*Detection of one or more chromosomal aberrations resp. of one or more somatic mutations with an allele frequency of ≥2% (or ≥4% for X-linked gene mutations in males), including mutations in specific genes (see Molecular Genetics, Table 1):
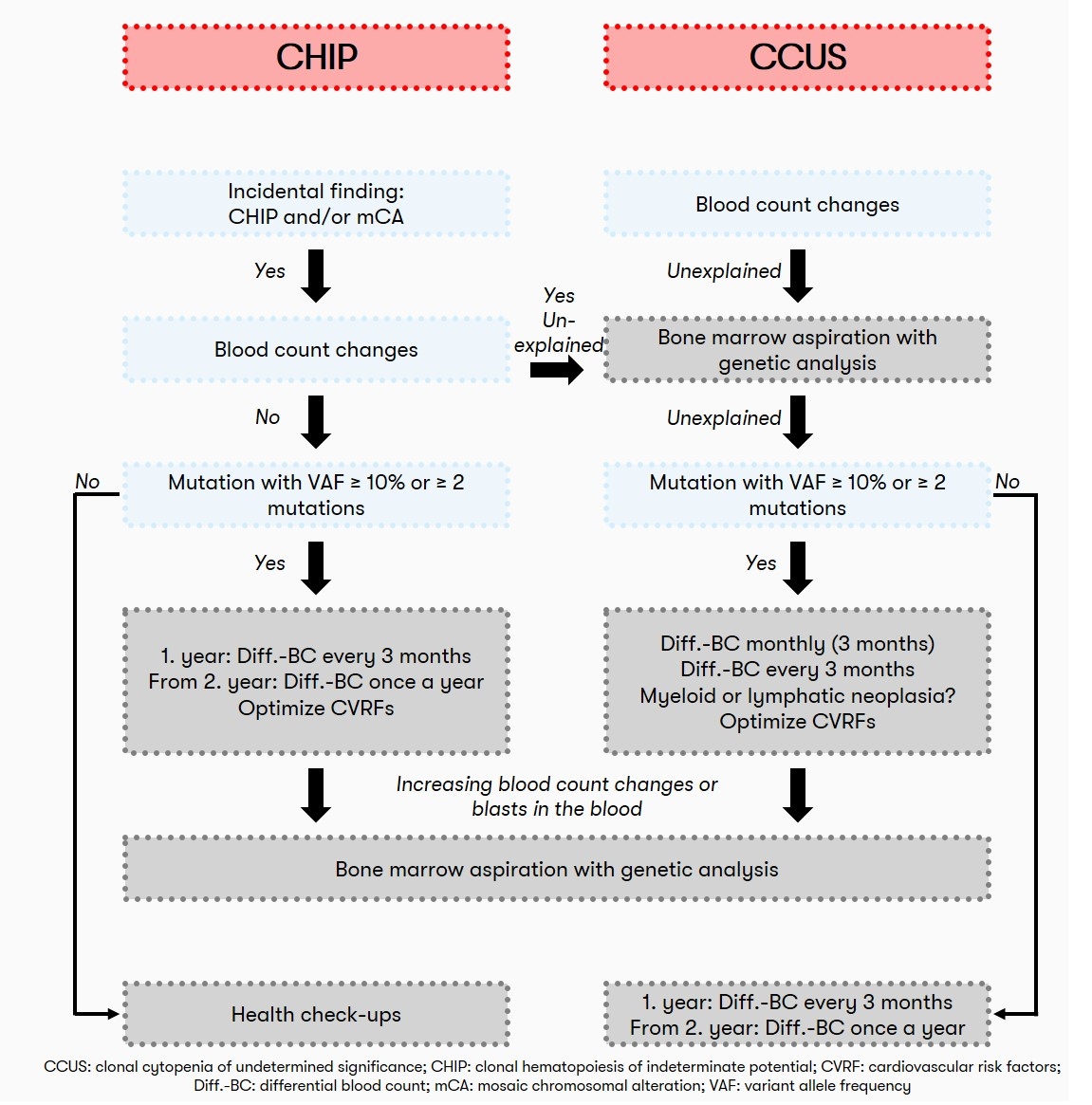
Differentiation of CCUS from CHIP and MDS
Clonal cytopenia of indeterminate significance should be distinguished from clonal hematopoiesis of indeterminate potential (CHIP), in which acquired molecular genetic alterations are detectable but cytopenia is not present.
CCUS and myelodysplastic neoplasm (MDS) have cytopenia(s) and clonal alterations in common, but in CCUS the diagnostic criteria for classification according to WHO 2022 into the MDS subgroups "MDS with defining genetic alterations" or "MDS, morphologically defined" are not met.
Further information on MDS and MDS diagnostics can be found here.
Diagnostic methods in clonal cytopenia of undetermined significance
Prognosis in clonal cytopenia of undetermined significance
Predictive and prognostic value of clonal hematopoiesis in unclear cytopenias
Already the detection or exclusion of clonality has prognostic relevance and utility for differential diagnosis. If no mutation can be detected, the probability of diagnosing a myeloid neoplasia at the time of examination or during follow-up is low (high negative predictive value) (Malcovati et al. 2017, Shanmugam et al. 2019, Rossi et al. 2021).
Number, type, and clone size of detected alterations influence risk for hematologic neoplasia
Clonal hematopoiesis is associated with an increased likelihood that a hematologic neoplasia will exist or occur later in life. The positive predictive value for the presence of myeloid neoplasia can be maximized when a higher allele frequency (≥10% or ≥20%) and/or the presence of ≥2 gene mutations are applied as criteria (Malcovati et al. 2017, Shanmugam et al. 2019, Zheng et al. 2019, Galli et al. 2021). Certain mutations are also predictive of MDS - these include, in particular, mutations in spliceosome factors (SF3B1, SRSF2, U2AF1, ZRSR2) (Malcovati et al. 2017, Shanmugam et al. 2019, Galli et al. 2021) or RUNX1 and JAK2 (Malcovati et al. 2017). If DNMT3A, TET2, or ASXL1 mutations are present in combination with at least one other mutation, this mutation pattern is also predictive of MDS (Malcovati et al. 2017, Galli et al. 2021). A study on the significance of clonal hematopoiesis in individuals ≥80 years of age showed that overall survival of individuals with unclear cytopenia in combination with specific mutations is not different from patients with diagnosed myeloid neoplasia (Rossi et al. 2021).
The study by Malcovati et al. (2017) showed a risk of progression to myeloid neoplasia of approximately 20% per year for patients with cytopenia and the specific mutation patterns mentioned above. Detection of other mutations or mutational constellations in persistent cytopenia was associated with a lower risk of progression to myeloid neoplasia (progression risk of approximately 10% per year). Galli et al (2021) found an association of mutations in DTA genes (DNMT3A, TET2, ASXL1) and SF3B1 or TP53 and additional mutations with myelodysplasias already at an allele frequency of <20%. In contrast, mutation patterns involving SRSF2, isolated or multiple DTA mutations, or other multiple mutations were associated with dysplasias only at allele frequencies >30%.
The "Clonal Hematopoiesis Risk Score (CHRS)" provides a framework for the prognostic assessment of the risk of progression to a hematological neoplasia. This examines various variables and classifies them into low, intermediate and high risk groups. In the CHRS, the sole presence of a DNMT3A mutation is assessed as a prognostically favorable factor. Prognostically unfavorable factors, on the other hand, are as follows (Weeks et al. 2023):
- Defined high-risk mutation (SRSF2, SF3B1, ZRSR2, IDH1, IDH2, FLT3, RUNX1, JAK2 and TP53).
- Detection of multiple mutations
- Clone size of at least 20% variant allele frequency (VAF)
- Red cell distribution width (RDW) of ≥15%
- Macrocytosis defined as mean red cell volume (MCV) of ≥100 fl
- Presence of cytopenia (CCUS vs. CHIP)
- Age ≥65 years
Recommendation in clonal cytopenia of undetermined significance
If the individual risk of progression (cf. also Prognosis) is strongly increased, especially in the case of one or more mutations with an allele frequency ≥10% or the presence of ≥2 mutations, quarterly monitoring by differential blood count is recommended with regard to progression to a hematologic neoplasm. In cases with a lower risk of progression, the recommendation is quarterly monitoring in the first year and annual monitoring from the second year onwards (Haferlach & Heuser 2022, see also Figure 2).
Status: March 2024